CARP: Continuous, Adaptive, Range Partitioner
Ingestion of data generated by high-performance scientific applications continues to stress available storage resources. Efficient range-based analyses on this data can be enabled by reordering it on attributes of interest, but require expensive post-processing sorts to realize the query benefits of reordering. In-situ indexing techniques, while write-efficient, are orders of magnitude slower at range queries than sorted indices. Range queries are necessary for analyzing continuous physical attributes and tracking phenomena such as energy bands and wave fronts.
CARP is a scalable data partitioner designed for high-throughput ingestion and range query indexing. With CARP, data is reordered in-situ as it is streamed to storage during application I/O. Motivated by our findings that real application distributions tend to be highly skewed and dynamic, CARP dynamically discovers and adapts its data partitions to track these characteristics. Our experiments (figure 1) show that CARP query performance is comparable to that of a full sort, the gold standard for range queries. On the write path (figure 2), CARP incurs no ingestion overhead, making it 5X faster than prior work.
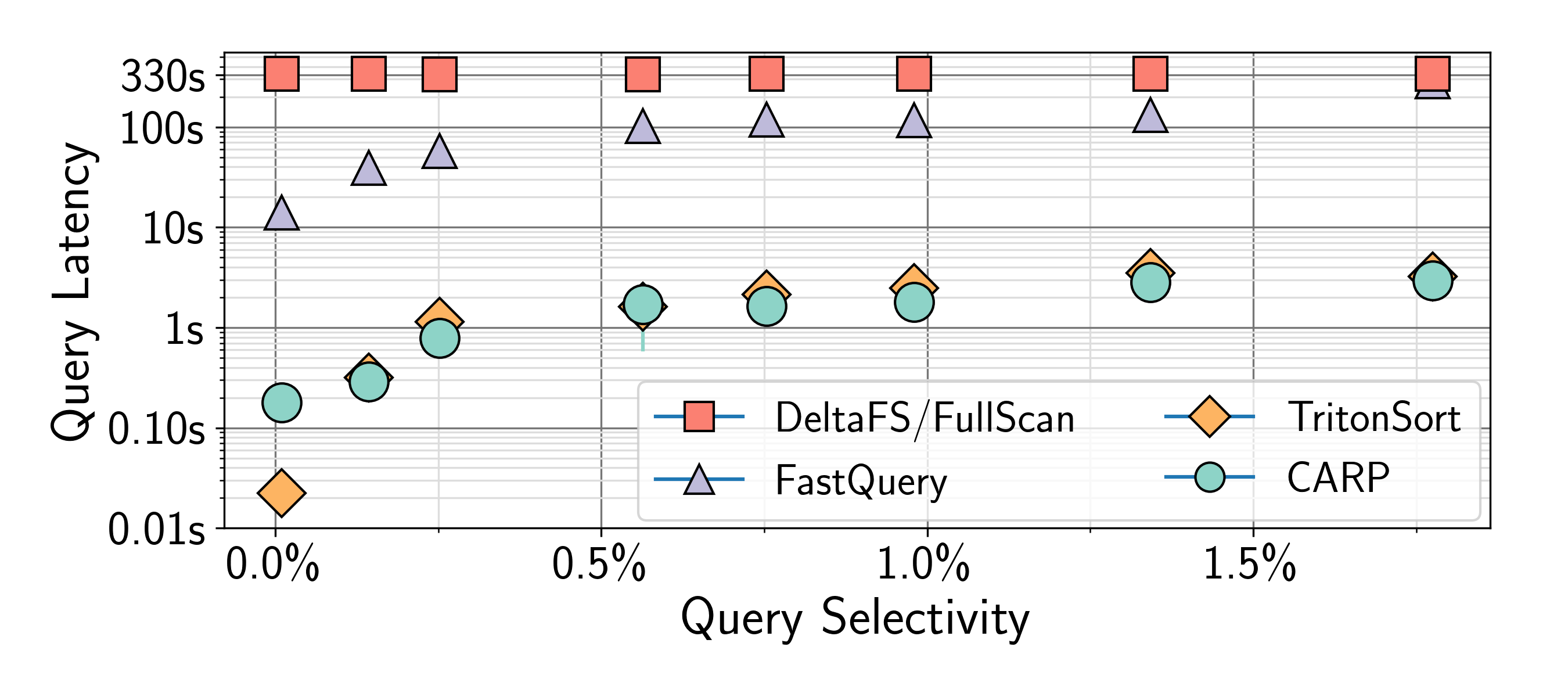
Fig 1. CARP query latency is comparable to a full sort (TritonSort),
and two orders of magnitude faster than bitmap indices (FastQuery).
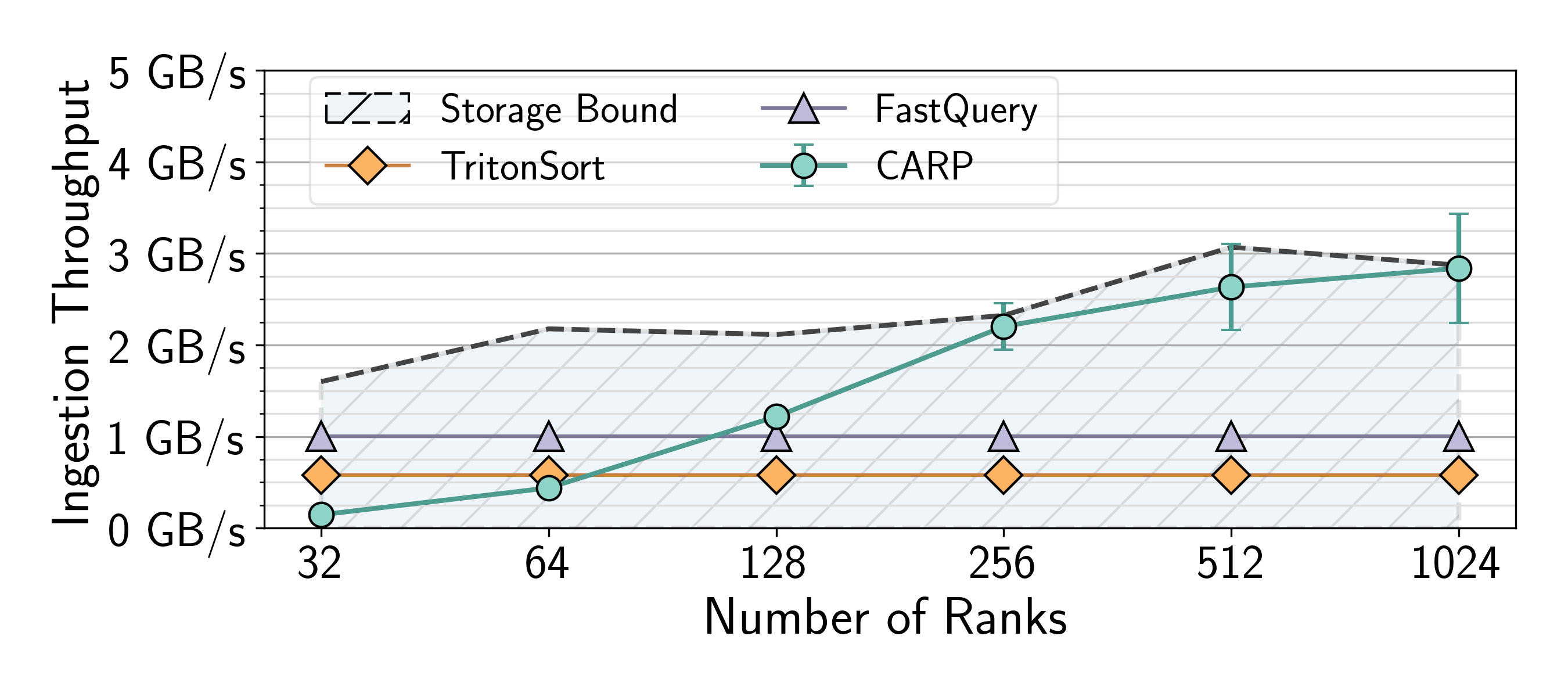
Fig 2. CARP ingests data at the maximum storage layer bandwidth,
3X faster than FastQuery and 5X faster than TritonSort.
People
FACULTY
George Amvrosiadis
Chuck Cranor
GRAD STUDENTS
ALUMNI
Qing Zheng
PARTNERS
Los Alamos National Laboratory: Gary Grider, Brad Settlemyer, Fan Guo
Publications
- CARP: Range Query-Optimized Indexing for Streaming Data. Ankush Jain, Charles D. Cranor, Qing Zheng, Bradley W. Settlemeyer, George Amvrosiadis, Gary Grider. SC24, November 17-22, 2024, Atlanta, Georgia, USA.
Abstract / PDF [1M]
Acknowledgements
We thank the members and companies of the PDL Consortium: Bloomberg LP, Datadog, Google, Intel Corporation, Jane Street, LayerZero Labs, Meta, Microsoft Research, Oracle Corporation, Oracle Cloud Infrastructure, Pure Storage, Salesforce, Samsung Semiconductor Inc., Uber, and Western Digital for their interest, insights, feedback, and support.